Voxelwise Encoding Bids
Published:
An example workflow for voxel-wise encoding models using a BIDS app
This shows how to (for a BIDS compliant dataset) extract features, save them in BIDS format, and run a BIDS app for voxel-wise encoding models. We are going to use this dataset.
Warning: Executing this notebook will download the full dataset.
!aws s3 sync --no-sign-request s3://openneuro.org/ds002322 ds002322-download/
Extracting a stimulus representation
The dataset in question consists of fMRI activity recorded of several participants while they listened to a reading of the first chapter of Lewis Carroll’s Alice in Wonderland. First we want to extract a stimulus representation that we can use - I chose a Mel spectrogram for demonstration. This small Python script extracts such a representation and saves it in a BIDS compliant format.
If you get an error that sndfile library was not found, you will need to use conda to install it.
import json
# these are the parameters for extracting a Mel spectrogram
# for computational ease in this example we want 1 sec segments of 31 Mel frequencies with a max frequency of * KHz
mel_params = {'n_mels': 31, 'sr': 16000, 'hop_length': 16000, 'n_fft': 16000, 'fmax': 8000}
with open('config.json', 'w+') as fl:
json.dump(mel_params, fl)
!git clone https://github.com/mjboos/audio2bidsstim/
!pip install -r audio2bidsstim/requirements.txt
!python audio2bidsstim/wav_files_to_bids_tsv.py ds002322-download/stimuli/DownTheRabbitHoleFinal_mono_exp120_NR16_pad.wav -c config.json
!ls -l
Now we must copy these files into the BIDS dataset directory according to these specifications. We are going to use the derivatives folder for the already preprocessed data.
!cp DownTheRabbitHoleFinal_mono_exp120_NR16_pad.tsv.gz ds002322-download/derivatives/task-alice_stim.tsv.gz
!cp DownTheRabbitHoleFinal_mono_exp120_NR16_pad.json ds002322-download/derivatives/sub-18/sub-18_task-alice_stim.json
And, lastly, because for this dataset the derivatives folder is missing timing information for the BOLD files - we are only interested in the TR - we have to copy that as well.
!cp ds002322-download/sub-18/sub-18_task-alice_bold.json ds002322-download/derivatives/sub-18/sub-18_task-alice_bold.json
Running the analysis
Now we’re all set and can run our encoding analysis. This analysis uses standard Ridge regression, and we’re going to specify some additional parameters here.
ridge_params = {'alphas': [1e-1, 1, 100, 1000], 'n_splits': 3, 'normalize': True}
# and for lagging the stimulus as well - we want to include 6 sec stimulus segments to predict fMRI
lagging_params = {'lag_time': 6}
with open('encoding_config.json', 'w+') as fl:
json.dump(ridge_params, fl)
with open('lagging_config.json', 'w+') as fl:
json.dump(lagging_params, fl)
Now we just need this BIDS app for running the analysis. Running this cell will fit voxel-wise encoding models, which right now need about 8 Gig of RAM.
Using Docker to run the voxelwise-encoding BIDS app
You can use Docker to build/get an image that already includes all libraries:
!git clone https://github.com/mjboos/voxelwiseencoding
!mkdir output
# we need to mount a config folder for our json files
!mkdir config
!cp *config.json config/
!docker run -i --rm -v ds002322-download/derivatives:bids_dataset/:ro -v config/:/config:ro -v output/:/output mjboos/voxelwiseencoding /bids_dataset /output --task alice --skip_bids_validator --participant_label 18 --preprocessing-config /config/lagging_config.json --encoding-config /config/encoding_config.json --detrend --standardize zscore
Alternative: run the module directly
Alternatively you can install the required libraries directly and run the Python script yourself.
!git clone https://github.com/mjboos/voxelwiseencoding
!pip install -r voxelwiseencoding/requirements.txt
!mkdir output
!python voxelwiseencoding/run.py ds002322-download/derivatives output --task alice --skip_bids_validator --participant_label 18 --preprocessing-config lagging_config.json --encoding-config encoding_config.json --detrend --standardize zscore
Now we’ll have some ridge regressions saved in output, as well as scores saved as a Nifti file - which we can visualize. First we load the scores - we have one volume containing the scores per fold - and average them and then plot them via Nilearn.
from nilearn.image import mean_img
mean_scores = mean_img('output/sub-18_task-alice_scores.nii.gz')
from nilearn import plotting
plotting.plot_stat_map(mean_scores, threshold=0.1)
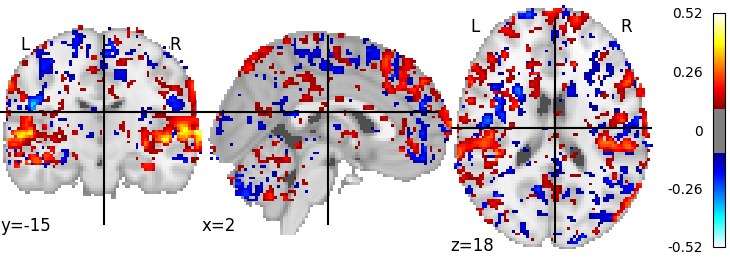
And voilà, we see that we can predict activity in the auditory areas.
You can download this notebook here.
