Unsupervised typo finder with dirty_cat
Published:
Finding misspelled names with dirty_cat and unsupervised learning
As a data scientist one often wants to group or analyze data conditional on a categorical variable. However, outside the world of neatly curated data sets, I often encounter the case that there can be slight misspellings in the category names: This happens when, for example, data input should use a drop down menu, but users are forced to input the category name by hand. Misspellings happen and analyzing the resulting data using a simple GROUP BY is not possible anymore.
This problem is however the perfect use case of unsupervised learning, a category of various statical methods that find structure in data without providing explicit labels/categories of the data a-priori. Specifically clustering of the distance between strings can be used to find clusters of strings that are similar to each other (e.g. differ only by a misspelling) and hence gives us an easy tool to flag potentially misspelled category names in an unsupervised manner.
I want to use this problem to showcase a nice library for “dirty data”, called dirty_cat. dirty_cat can do many more things than solve this specific problem (and there are of course many different tools that can compute string distances as well), but I found this one easy to use, well maintained, and with a clear API suitable to this problem.
An example
Imagine the following example: As a data scientist, our job is to analyze the data from a hospital ward. We notice that most of the cases involve the prescription of one of three different medications - “Contrivan”, “Genericon”, or “Zipholan”. However, data entry is manual and - either because the prescribing doctor’s handwriting was hard to decipher or due to mistakes during data input - there are multiple spelling mistakes for these three medications.
Let’s generate some example data that demonstrate this.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
def generate_example_data(examples, entries_per_example, prob_mistake_per_letter):
"""Helper function to generate data consisting of multiple entries per example.
Characters are misspelled with probability `prob_mistake_per_letter`"""
import string
data = []
for example, n_ex in zip(examples, entries_per_example):
len_ex = len(example)
# generate a 2D array of chars of size (n_ex, len_ex)
str_as_list = np.array([list(example)]*n_ex)
# randomly choose which characters are misspelled
idxes = np.where(np.random.random(len(example[0])*n_ex) < prob_mistake_per_letter)[0]
# and randomly pick with which character to replace
replacements = [string.ascii_lowercase[i] for i in np.random.choice(np.arange(26), len(idxes)).astype(int)]
# introduce spelling mistakes at right examples and right char locations per example
str_as_list[idxes // len_ex, idxes % len_ex] = replacements
# go back to 1d array of strings
data.append(np.ascontiguousarray(str_as_list).view(f'U{len_ex}').ravel())
return np.concatenate(data)
np.random.seed(123)
# our three medication names
medications = ["Contrivan", "Genericon", "Zipholan"]
entries_per_medications = [500, 100, 1500]
# 5% probability of a typo per letter
prob_mistake_per_letter = 0.05
data = generate_example_data(medications, entries_per_medications, prob_mistake_per_letter)
# we extract the unique medication names in the data & how often they appear
unique_examples, counts = np.unique(data, return_counts=True)
# and build a series out of them
ex_series = pd.Series(counts, index=unique_examples)
Visualize the data
ex_series.plot.bar(figsize=(20, 10))
_=plt.xlabel('Medication name')
_=plt.ylabel('Counts')
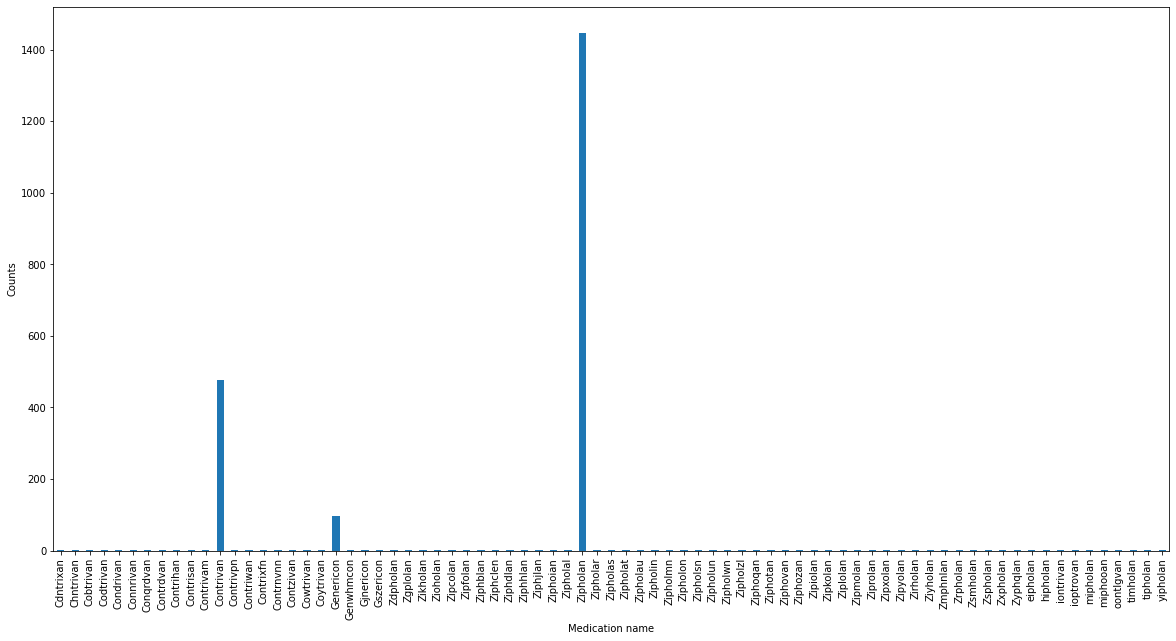
We can now see clearly the structure of the data: The three original medications are the most common ones, however there are many spelling mistakes and hence many slight variations of the names of the original medications.
The idea is to use the fact that the string-distance of each misspelled medication name will be closest to either the correctly or incorrectly spelled orginal medication name - and therefore form clusters.
from dirty_cat import SimilarityEncoder
from sklearn.preprocessing import minmax_scale
from scipy.spatial.distance import squareform
simenc = SimilarityEncoder(similarity="jaro-winkler")
# transform sorted entity names to similarities
transf = simenc.fit_transform(ex_series.index.values[:, None])
corr_dist = minmax_scale(-transf)
dense_distance = squareform(corr_dist, checks=False)
We can visualize the pair-wise distance between all medication names
Below I use a heatmap to visualize the pairwise-distance between medication names. A darker color means that two medication names are closer together (are more similar), a lighter color means a larger distance. It’s clear that we are dealing with three clusters - the original medication names and their misspellings that cluster around them.
import seaborn as sns
fig, axes = plt.subplots(1, 1, figsize=(12, 12))
sns.heatmap(corr_dist, yticklabels=ex_series.index, xticklabels=ex_series.index, ax=axes)
<AxesSubplot:>
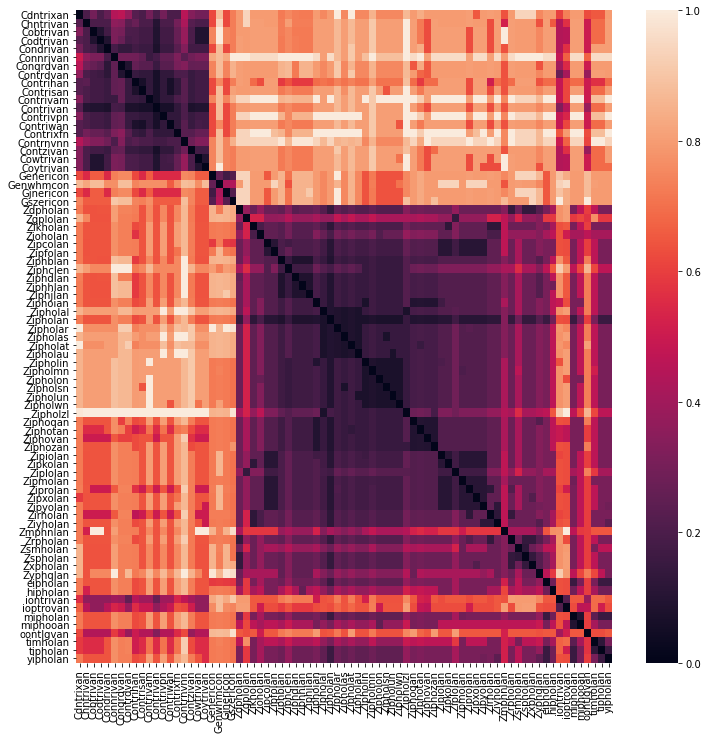
Clustering to suggest corrections of misspelled names
The “strictness” parameter will need some adjustment depending on the data you have - it is the cutoff in the hierarchical clustering and will differ depending on how many spelling errors you expect. It can in principle be set automatically via any clustering quality criterion, but I’d advise checking whether it works in practice for your data (this assumes that the prevalence of spelling mistakes stays roughly the same across time).
from scipy.cluster.hierarchy import linkage, fcluster
Z = linkage(dense_distance, "average", optimal_ordering=True)
clstrs = fcluster(Z, 0.7, criterion="distance")
replace_dict = {}
for cluster in np.unique(clstrs):
# get the most common medication name in each cluster
correct_spelling, misspellings = np.array_split(
ex_series.loc[clstrs==cluster].sort_values(ascending=False).index.values, [1,])
print(f"Cluster {cluster} with most common (likely correct) spelling {correct_spelling[0]} and likely misspellings {misspellings}.")
# now add misspellings to dict for replacing later
for misspelled in misspellings:
replace_dict[misspelled] = correct_spelling[0]
Cluster 1 with most common (likely correct) spelling Zipholan and likely misspellings ['Zipfolan' 'yipholan' 'Ziphotan' 'miphooan' 'Ziphozan' 'Zipiolan'
'Zipkolan' 'Ziplolan' 'Zipmolan' 'Ziprolan' 'Zipxolan' 'Zipyolan'
'Zirholan' 'Ziyholan' 'timholan' 'Zrpholan' 'Ziphovan' 'Zsmholan'
'Zspholan' 'Zxpholan' 'Zyphqlan' 'eipholan' 'hipholan' 'mipholan'
'tipholan' 'Zmphnlan' 'Zdpholan' 'Zgplolan' 'Zipholal' 'Zikholan'
'Zioholan' 'Zipcolan' 'Ziphblan' 'Ziphclen' 'Ziphdlan' 'Ziphhlan'
'Ziphjlan' 'Ziphoian' 'Zipholar' 'Zipholzl' 'Zipholas' 'Zipholat'
'Zipholau' 'Zipholin' 'Zipholmn' 'Zipholon' 'Zipholsn' 'Zipholun'
'Zipholwn' 'Ziphoqan'].
Cluster 2 with most common (likely correct) spelling Contrivan and likely misspellings ['Contzivan' 'Chntrivan' 'ioptrovan' 'iontrivan' 'Coytrivan' 'Cowtrivan'
'Contrnvnn' 'Contrixfn' 'Contriwan' 'Contrivpn' 'Cdntrixan' 'Contrivam'
'Contrisan' 'Contrihan' 'Contrdvan' 'Conqrdvan' 'Connrivan' 'Condrivan'
'Codtrivan' 'Cobtrivan' 'oontlgvan'].
Cluster 3 with most common (likely correct) spelling Genericon and likely misspellings ['Genwhmcon' 'Gjnericon' 'Gszericon'].
Let’s see how it works when we apply the spelling correction:
# we just replace each medication in data by its correct spelling if it's in replacement_dict
# if not keep the original spelling
corrected_data = [replace_dict.get(dt, dt) for dt in data]
corrected_unique_examples, corrected_counts = np.unique(corrected_data, return_counts=True)
corrected_series = pd.Series(corrected_counts, index=corrected_unique_examples)
And we can do the same plot as before, just with the corrected data.
corrected_series.plot.bar(figsize=(20, 10))
_=plt.xlabel('Medication name')
_=plt.ylabel('Counts')
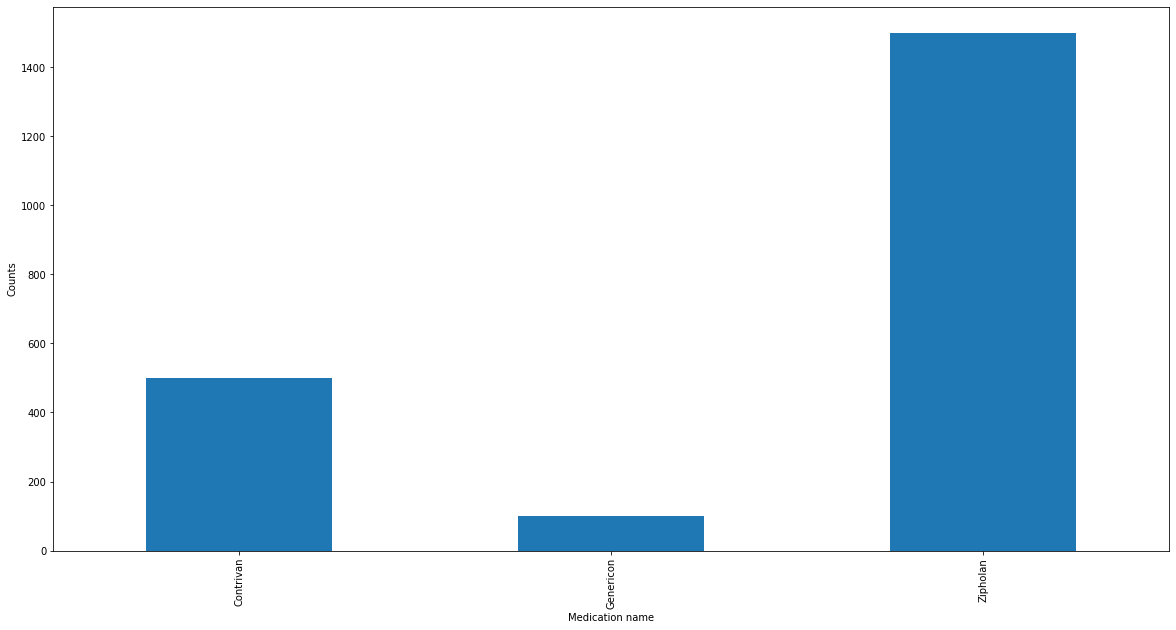
Luckily (and by choosing a simple example and the right clustering cutoff parameter) we correct all spelling mistakes. Hurrah!
What I hope this showcases is, how easy it is to implement an adhoc spelling mistake detector using dirty_cat to mitigate a problem that is all too common in the real world. In general, the best use for such a method is to give you an easy way to flag potential misspelling in an unsupervised fashion, not necessarily to automatically correct them.
